Substructure Similarity Search In Graph Databases
Introduction: Bioinformatics and chem-informatics applications involve query-based search in massive, complex structural data. Even with a graph index, such search can encounter challenges because it is often too restrictive to search for an exact match of an index entry. Efficient similarity search of complex structures becomes a vital operation.
Let’s examine a simple example.
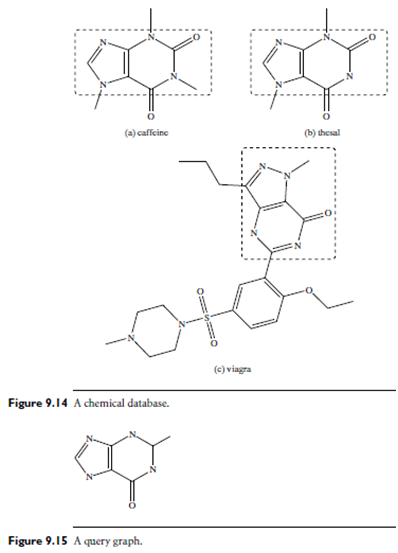 Example: Similarity search of chemical structures. Figure 9.14 is a chemical data set with three molecules. Figure 9.15 shows a substructure query. Obviously, no match exists for this query graph. If we relax the query by taking out one edge, then caffeine and thesal in Figure 9.14(a) and 9.14(b) will be good matches. If we relax the query further, the structure in Figure 9.14(c) could also be an answer.
Example: Similarity search of chemical structures. Figure 9.14 is a chemical data set with three molecules. Figure 9.15 shows a substructure query. Obviously, no match exists for this query graph. If we relax the query by taking out one edge, then caffeine and thesal in Figure 9.14(a) and 9.14(b) will be good matches. If we relax the query further, the structure in Figure 9.14(c) could also be an answer.
A naïve solution to this similarity search problem is to form a set of subgraph queries with one or more edge deletions and then use the exact substructure search. This does not work when the number of deletions is more than 1. For example, if we allow three edges to be deleted in a 20-edge query graph, it may generate (20 /3) = 1140 substructure queries, which are too expensive to check.
A feature-based structural filtering algorithm, called Grafil (Graph Similarity Filtering), was developed to filter graphs efficiently in large-scale graph databases. Grafil models each query graph as a set of features and transforms the edge deletions into “feature misses” in the query graph. It is shown that using too many features will not leverage the filtering performance. Therefore, a multi-filter composition strategy is developed, where each filter uses a distinct and complementary subset of the features. The filters are constructed by a hierarchical, one-dimensional clustering algorithm that groups features with similar selectivity into a feature set. Experiments have shown that the multi-filter strategy can significantly improve performance for a moderate relaxation ratio.